With the help of a quarter-million video game players, Princeton researchers have created and shared detailed maps of more than 1,000 neurons — and they’re just getting started.
“Working with Eyewirers around the world, we've made a digital museum that shows off the intricate beauty of the retina's neural circuits,” said Sebastian Seung, the Evnin Professor in Neuroscience and a professor of computer science and the Princeton Neuroscience Institute (PNI). The related paper appears today in the journal Cell.
Seung is unveiling the Eyewire Museum, an interactive archive of neurons available to the general public and neuroscientists around the world, including the hundreds of researchers involved in the federal Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Initiative.
“This interactive viewer is a huge asset for these larger collaborations, especially among people who are not physically in the same lab,” said Amy Robinson Sterling, a crowdsourcing specialist with PNI and the executive director of Eyewire, the online gaming platform for the citizen scientists who have created this data set.
“This museum is something like a brain atlas,” said Alexander Bae, a graduate student in electrical engineering and one of four co-first authors on the paper. “Previous brain atlases didn’t have a function where you could visualize by individual cell, or a subset of cells, and interact with them. Another novelty: Not only do we have the morphology of each cell, but we also have the functional data, too.”
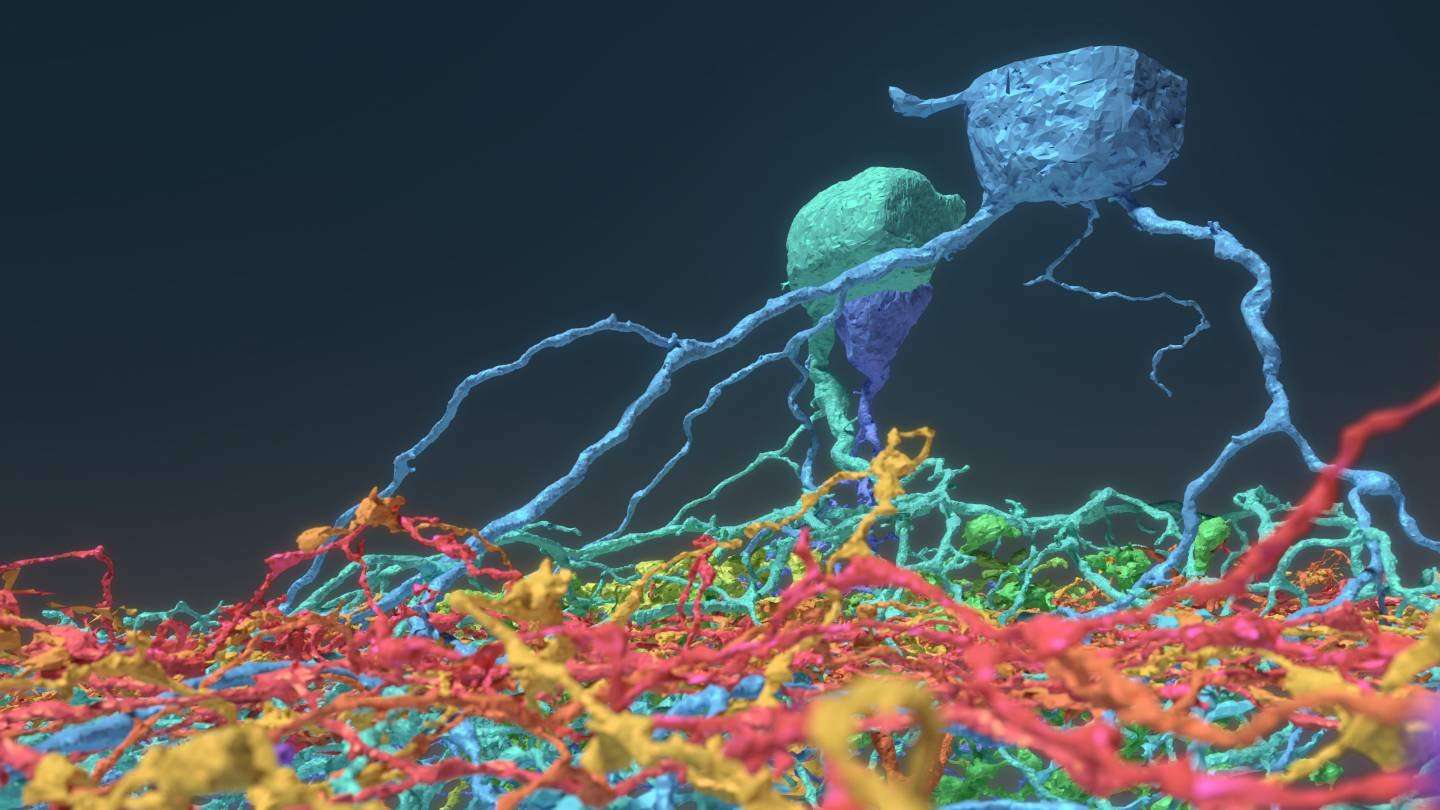
Eyewire gamers mapped this synapse between a ganglion neuron (blue) and a starburst amacrine cell (yellow).
The neural maps were developed by Eyewirers, members of an online community of video game players who have devoted hundreds of thousands of hours to painstakingly piecing together these neural cells, using data from a mouse retina gathered in 2009.
Eyewire pairs machine learning with gamers who trace the twisting and branching paths of each neuron. Humans are better at visually identifying the patterns of neurons, so every player’s moves are recorded and checked against each other by advanced players and Eyewire staffers, as well as by software that is improving its own pattern recognition skills.
Since Eyewire’s launch in 2012, more than 265,000 people have signed onto the game, and they’ve collectively colored in more than 10 million 3-D “cubes,” resulting in the mapping of more than 3,000 neural cells, of which about a thousand are displayed in the museum.
Each cube is a tiny subset of a single cell, about 4.5 microns across, so a 10-by-10 block of cubes would be the width of a human hair. Every cell is reviewed by between 5 and 25 gamers before it is accepted into the system as complete.
“Back in the early years it took weeks to finish a single cell,” said Sterling. “Now players complete multiple neurons per day.” The Eyewire user experience stays focused on the larger mission — “For science!” is a common refrain — but it also replicates a typical gaming environment, with achievement badges, a chat feature to connect with other players and technical support, and the ability to unlock privileges with increasing skill. “Our top players are online all the time — easily 30 hours a week,” Sterling said.
Dedicated Eyewirers have also contributed in other ways, including donating the swag that gamers win during competitions and writing program extensions “to make game play more efficient and more fun,” said Sterling, including profile histories, maps of player activity, a top 100 leaderboard and ever-increasing levels of customizability.
“The community has really been the driving force behind why Eyewire has been successful,” Sterling said. “You come in, and you’re not alone. Right now, there are 43 people online. Some of them will be admins from Boston or Princeton, but most are just playing — now it’s 46.”
Play Video: 6 New Neurons Discovered by Eyewirers These six new types of neurons were first mapped by gamers playing Eyewire, a project of Princeton's Seung Lab, and they are now available to explore in the new interactive cell visualizer at museum.eyewire.org.
With 100 billion neurons linked together via trillions of connections, the brain is immeasurably complex, and neuroscientists are still assembling its “parts list,” said Nicholas Turner, a graduate student in computer science and another of the co-first authors. “If you know what parts make up the machine you’re trying to break apart, you’re set to figure out how it all works,” he said.
The researchers have started by tackling Eyewire-mapped ganglion cells from the retina of a mouse. “The retina doesn't just sense light,” Seung said. “Neural circuits in the retina perform the first steps of visual perception.”
The retina grows from the same embryonic tissue as the brain, and while much simpler than the brain, it is still surprisingly complex, Turner said. “Hammering out these details is a really valuable effort,” he said, “showing the depth and complexity that exists in circuits that we naively believe are simple.”
The researchers’ fundamental question is identifying exactly how the retina works, said Bae. “In our case, we focus on the structural morphology of the retinal ganglion cells.”
“Why the ganglion cells of the eye?” asked Shang Mu, an associate research scholar in PNI and fellow first author. “Because they’re the connection between the retina and the brain. They’re the only cell class that go back into the brain.” Different types of ganglion cells are known to compute different types of visual features, which is one reason the museum has linked shape to functional data.
Members of the Seung Lab, from left: Sitting: Alexander Bae, Thomas Macrina, Sergiy Popovych, Jingpeng Wu; Standing: Nicholas Turner, Sven Dorkenwald, Chris Jordan, Davit Buniatyan, Zhen Jia, Sebastian Seung, William Silversmith, Ran Lu, Kisuk Lee, Kyle Luther, Nico Kemnitz, William Wong, Shang Mu, Alyssa Wilson, Maysoun Husseini.
Using Eyewire-produced maps of 396 ganglion cells, the researchers in Seung’s lab successfully classified these cells more thoroughly than has ever been done before.
“The number of different cell types was a surprise,” said Mu. “Just a few years ago, people thought there were only 15 to 20 ganglion cell types, but we found more than 35 — we estimate between 35 and 50 types.”
Of those, six appear to be novel, in that the researchers could not find any matching descriptions in a literature search.
"This does represent a significant contribution," said Marla Feller, professor and head of the Division of Neurobiology at the University of California-Berkeley's Department of Molecular and Cell Biology & Helen Wills Neuroscience Institute, who was not involved in the research. "Establishing a relationship between the structure and function of a neuron is a primary goal of neurobiologists. However, structures are too small and dense to be able to make sense of with the human eye or even with an optical microscope. By completely reconstructing neighboring neurons of the same type within one piece of the retina, we can now examine their spatial relationships."
A brief scroll through the digital museum reveals just how remarkably flat the neurons are — nearly all of the branching takes place along a two-dimensional plane. Seung’s team discovered that different cells grow along different planes, with some reaching high above the nucleus before branching out, while others spread out close to the nucleus. Their resulting diagrams resemble a rainforest, with ground cover, an understory, a canopy and an emergent layer overtopping the rest.
All of these are subdivisions of the inner plexiform layer, one of the five previously recognized layers of the retina. The researchers also identified a “density conservation principle” that they used to distinguish types of neurons.
One of the biggest surprises of the research project has been the extraordinary richness of the original sample, said Seung. “There’s a little sliver of a mouse retina, and almost 10 years later, we’re still learning things from it.”
“Digital museum of retinal ganglion cells with dense anatomy and physiology,” by Alexander Bae, Shang Mu, Jinseop Kim, Nicholas Turner, Ignacio Tartavull, Nico Kemnitz, Chris Jordan, Alex Norton, William Silversmith, Rachel Prentki, Marissa Sorek, Celia David, Devon Jones, Doug Bland, Amy Sterling, Jungman Park, Kevin Briggman, Sebastian Seung and the Eyewirers, was published May 17 in the journal Cell with DOI 10.1016/j.cell.2018.04.040. The research was supported by the Gatsby Charitable Foundation, National Institute of Health-National Institute of Neurological Disorders and Stroke (U01NS090562 and 5R01NS076467), Defense Advanced Research Projects Agency (HR0011-14-2- 0004), Army Research Office (W911NF-12-1-0594), Intelligence Advanced Research Projects Activity (D16PC00005), KT Corporation, Amazon Web Services Research Grants, Korea Brain Research Institute (2231-415) and Korea National Research Foundation Brain Research Program (2017M3C7A1048086).
yaboibowie on May 20th, 2018 at 14:13 UTC »
Hey, this is eyewire! I played a lot of citizen science game a couple years back. I was into eyewire for a time and it was super fun. You are basically filling in a cube using a cross sectional piece trying to find which way the neuron goes. You have to rely on how something visually looks in the 3d model to see if it is in fact the same neuron as the one that you were instructed to originally trace. I would really recommend it as you can learn it very quickly and it takes very little time commitment.
Could_be_cubing on May 20th, 2018 at 13:37 UTC »
I wonder if you could use this data to train a neural network or some other ML algorithm to automate the process of creating the volumetric model from the microscope images. 1000 seems like a large enough sample size to get an accurate result, and more could be crowdsourced.
^(and yes the irony of using neural networks to map neurons is not lost on me)
mvea on May 20th, 2018 at 10:50 UTC »
The title of the post is a copy and paste from the title, subtitle and first paragraph of the linked academic press release here :
Journal Reference:
Digital Museum of Retinal Ganglion Cells with Dense Anatomy and Physiology
Bae, J. Alexander et al.
Cell , Volume 173 , Issue 5 , 1293 - 1306.e19
DOI: https://doi.org/10.1016/j.cell.2018.04.040
Link: https://www.cell.com/cell/fulltext/S0092-8674(18)30572-5
Highlights
•A digital “museum” provides dense anatomy and physiology of retinal ganglion cells •The inner plexiform layer divides into four sublaminae defined by anatomical criteria •The aggregate neurite density of a ganglion cell type is approximately uniform •Inner marginal ganglion cells exhibit significantly more sustained visual responses
Summary
When 3D electron microscopy and calcium imaging are used to investigate the structure and function of neural circuits, the resulting datasets pose new challenges of visualization and interpretation. Here, we present a new kind of digital resource that encompasses almost 400 ganglion cells from a single patch of mouse retina. An online “museum” provides a 3D interactive view of each cell’s anatomy, as well as graphs of its visual responses. The resource reveals two aspects of the retina’s inner plexiform layer: an arbor segregation principle governing structure along the light axis and a density conservation principle governing structure in the tangential plane. Structure is related to visual function; ganglion cells with arbors near the layer of ganglion cell somas are more sustained in their visual responses on average. Our methods are potentially applicable to dense maps of neuronal anatomy and physiology in other parts of the nervous system.