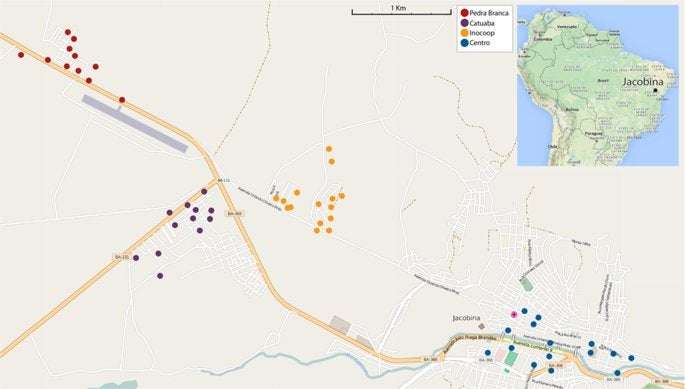
Jacobina, in the state of Bahia, Brazil, is a moderately sized city of ~75,000 inhabitants located at coordinates 11°10′51″S, 40°31′04″W (Fig. 1). Jacobina is surrounded for several kilometers in all directions by caatinga, a dry ecological biome in which Ae. aegypti cannot breed, making Jacobina an island for this mosquito.
Figure 1 Map of Jacobina. Ovitraps where samples were collected are indicated with colored dots, coded by neighborhood. Releases were made in the neighborhoods of Pedra Branca, Catuaba, and Inocoop but never in the Centro area. © OpenStreetMap contributors. Full size image
The rearing facility for the release strain is located at the Biofabrica Moscamed Brasil in Juazeiro, some 200 kilometers north of Jacobina. Mass rearing and sexing are described in Harris et al.7. Weekly, male pupae were transported to Jacobina and held in a local facility for one week to allow eclosion before release; approximately 450 thousand OX513A males were released each week beginning in June 2013 and continued through September 20156. Releases were made in the Pedra Branca, Catuaba, and Inocoop neighborhoods, but never in Centro. Oviposition traps were sampled weekly in the localities indicated in Fig. 1. Eggs were hatched and the frequencies of fluorescent and wild type larvae recorded; see Garzeira et al.6 for details of proportion fluorescent and wildtype at each time point. Fourth instar larvae of each type were placed in ~80% ethanol and brought to Yale University of genotyping. Further data on the effect of releases in Jacobina can be found in Graziera et al.6.
We used a custom developed Affymetrix SNP chip for genotyping8. Approximately 200 ng of genomic DNA from individual mosquitoes were placed in 95 wells of a 96 well plate, with one distilled water control. Plates were sent to the Functional Genomics Core at the University of North Carolina, Chapel Hill, for hybridization and production of data files sent to Yale University. We used the R package SNPolisher v1.4 (Afffymetrix, Santa Clara, CA) to generate and process genotype calls. While the SNP chip contains probes for about 27,000 well-validated biallelic SNPs passing tests for Mendelian inheritance and genotyping >98% of all samples8, 21,770 were polymorphic in our samples from Jacobina and genotyped in >98% of all individuals.
We genotyped samples taken from Centro and a combined Catuaba/Pedra Branca sample before releases began. Then, while the releases were continuing, we sampled all neighborhoods six, 12 and 27–30 months after releases began. The last sample at 27–30 months was a combined sample for three months included after the releases ceased at 27 months. Sample sizes are in Table 1. Except for the final combined 27–30 month sample, each sample analyzed after releases began were from egg traps exposed for a single week and larvae sampled from at least five traps in each neighborhood. The position of the traps remained the same throughout the study.
Table 1 Results of the “INTROGRESS” analysis as performed using the R package10. Full size table
To confirm our genetic analyses were accurate in detecting hybrids, we also genotyped 57 fluorescent larvae collected six months into the releases representing F 1 offspring between the release strain and the natural population.
We performed three types of analyses. First, to confirm that our panel of SNPs could discriminate between the release strain OX513A and the natural population before release, we performed a Principal Components Analysis (PCA) using the R package in LEA9. Second, the R package “introgress”10 was implemented designating OX513A and Jacobina before release (combined Centro, Catuaba, and Pedra Branca neighborhoods) as the two parental populations. Third, we performed an ADMIXTURE analysis as describe in11 and shown Fig. 2C. For this analysis we filtered to exclude tightly linked SNPs using the –indep option of PLINK12 resulting in a panel of 14,252 SNPs. Then, an ANOVA analysis followed by a post-hoc TukeyHSD test was used to test for statistical differences (confidence level 0.95) in the mean Q values between the populations and most importantly between the pre- and the post-release populations.
Figure 2 (A) Principal Components Analysis (PCA) on the OX513A release strain and three neighborhoods Jacobina (Centro and Catuaba/Pedra Branca) before releases began. (B) Hybrid index (h-index) as performed in INTROGRESS10. An index of 1.0 indicates the “pure” OX513A individuals, 0.0 indicates the “pure” Jacobina pre-release individuals. Individuals are organized by neighborhood indicated at bottom of the figure, then by collection date: pre-release, 6, 12 or 27–30 months post release. Fluorescence verified F1 hybrids are grouped and labeled as F1. The horizontal dashed line represents cutoff (h-index = 0.02) the maximum observed pre-release. (C) ADMIXTURE11 analysis of all individual genotypes. Proportion of each color for each individual represents the proportion of that individual’s ancestry attributable to the red (OX513A) or blue (Jacobina pre-release) cluster. Full size image
The dengue virus serotype 2 (DENV-2) strain tested was isolated during an epidemic in Brazil in 2010 from a patient in Santos, Brazil. The strain, designated ACS4613, was described in Cugola et al.14 and was kindly provided by the Evandro Chagas Institute in Belém, Pará.
The mosquito infection procedures are described in detail in Cost-da-Silva et al.15. Briefly, pre-mated five to seven day old females were blood-fed artificially using Glytube feeder (22). DENV-2 of ninth subculture (T9) or ZIKVBR of fourth subculture (T4) were mixed with human concentrated erythrocyte and inactivated blood serum to feed the females. DENV-2 and ZIKVBR final concentrations in the feeding solution were 1.7 × 1010 genome copies/ml and 2.2 × 106 plaque forming unit (pfu)/mL, respectively.
Engorged females from ROCK, OX513A and Jacobina strains were separated from non-engorged mosquitoes and maintained on 10% sucrose. Fourteen days post-blood meal (14 PBM), females were CO 2 anaesthetized and kept on ice. Individual mosquito bodies were separated from heads and frozen separately immediately on dry ice and stored at −80 °C. Total RNA was extracted using QIAamp Viral RNA Mini Kit (Qiagen). DENV-2 or ZIKVBR genomic copies were measured using one-step qRT-PCR method as described in (22). To generate DENV-2 standard curve, a 119-bp fragment from the ACS46 strain was amplified with D1-TS2 primers15 and was cloned into the pCR2.1 vector (Invitrogen). This plasmid was used to estimate the number of DENV copies for each sample. The thermocycler conditions for DENV-2 amplification were 48 °C for 30 min and 95 °C for 10 min; 45 cycles of 95 °C for 30 sec, 55 °C for 30 sec and 60 °C for 30 sec, and a melting curve step of 95 °C for 1 min, 60 °C for 30 sec and 95 °C for 1 min, with temperature ramping from 60 °C to 95 °C at 0.02 °C/sec.
Statistical analyses were performed to assess significant differences in viral levels (Kruskal-Wallis test followed by Dunn’s Multiple Comparison Test) or infection rates of heads or bodies (Fisher’s exact test) between the three mosquito strains. The program and procedures to perform the analyses were previously described15.
Mainfreed on September 16th, 2019 at 04:19 UTC »
Is also cool to add the fact that the Aedes aegypti isn't and endemic species from Brazil. Was brought here around the 1700, inside slave trading ships. In the 50s the Aedes population was almost eradicated, but the insect returned due the fact some neighboring countries weren't able to contain and eliminate the mosquito.
SelarDorr on September 16th, 2019 at 04:01 UTC »
I think the gravity of the article is misinterpreted here. This is not a report of good results. The genetics of the lab mosquitoes are not supposed to be transferred the wild population in high numbers, but evidently it is. There are now multiple genotypes in the wild, with unknown consequences, and the experiment has shown to not decrease the population of mosquitoes to the level expected.
The 85% you are citing is from a 2015 publication.
" it is clear from the data in Garziera et al.6 that the effectiveness of the release program began to break down after about 18 months, i.e., the population which had been greatly suppressed rebounded to nearly pre-release levels. "
" The three populations forming the tri-hybrid population now in Jacobina (Cuba/Mexico/Brazil) are genetically quite distinct (Extended Data Fig. E2), very likely resulting in a more robust population than the pre-release population due to hybrid vigor. "
questors on September 16th, 2019 at 03:51 UTC »
Aegypti, as indicated by the name, are from Africa and came to the rest of the world via modern transport. They have been extensively studied and found to not be a primary food source of birds or bats in the Western Hemisphere.